Protein Multimer Structure Prediction via PPI-guided Prompt Learning
Abstract
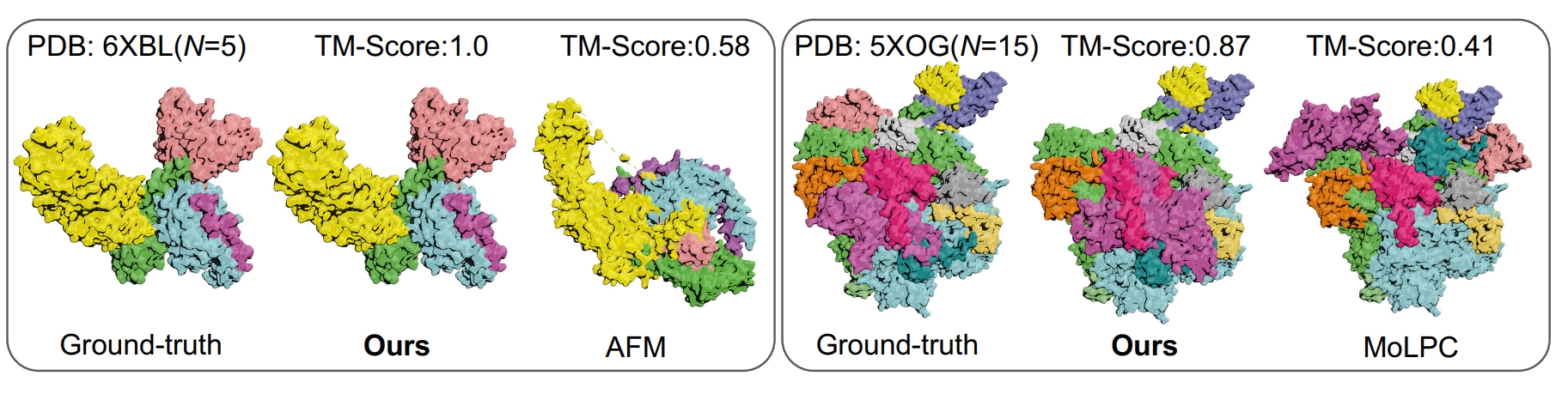
Understanding the 3D structures of protein multimers is crucial, as they play a vital role in regulating various cellular processes. It has been empirically confirmed that the multimer structure prediction (MSP) can be well handled in a step-wise assembly fashion using provided dimer structures and predicted protein-protein interactions (PPIs). However, due to the biological gap in the formation of dimers and larger multimers, directly applying PPI prediction techniques can often cause a poor generalization to the MSP task. To address this challenge, we aim to extend the PPI knowledge to multimers of different scales (i.e., chain numbers). Specifically, we propose PromptMSP, a pre-training and Prompt tuning framework for Multimer Structure Prediction. First, we tailor the source and target tasks for effective PPI knowledge learning and efficient inference, respectively. We design PPI-inspired prompt learning to narrow the gaps of two task formats and generalize the PPI knowledge to multimers of different scales. We utilize the meta-learning approach to learn a reliable initialization of the prompt model, enabling our prompting framework to effectively adapt to limited data for large-scale multimers. Empirically, we achieve both significant accuracy (RMSD and TM-Score) and efficiency improvements compared to advanced MSP models. For instance, when both methods utilize AlphaFold-Multimer to prepare dimers, PromptMSP achieves a 21.43% improvement in TM-Score with only 0.5% of the running time compared to the competitive MoLPC baseline.
Project members

Jia LI
Assistant Professor
Publications
Protein Multimer Structure Prediction via PPI-guided Prompt Learning. Ziqi Gao, Xiangguo Sun, Zijing Liu, Yu Li, Hong Cheng, and Jia Li.
Project Period
2024
Research Area
Data-driven AI
Keywords
docking path prediction, protein complex structure, prompt learning
