GALLOP: GPU Acceleration for Genomics Applications
Abstract
Graphics processors, or GPUs, have made high-performance computing inexpensive and widely accessible by packing hundreds of identical computing cores in a single chip. With the massive parallel processing power, GPUs have made their way into genomics applications through academic explorations as well as proprietary business solutions. Nevertheless, there lacks a large-scale, open, and systematic study of accelerating state-of-the-art genomic computing algorithms with the GPU. Therefore, we propose Gallop, an open-source software package that features new, GPU-accelerated algorithms for genomics applications.
Specifically, we are interested in four major computational tasks on genome data: (1) genome assembly, where short reads from an unknown DNA sequence are put together into a complete sequence; (2) sequence alignment, in which short reads are aligned to a reference sequence; (3) SNP (Single-Nucleotide Polymorphism) detection, through which the variation on a single nucleotide is identified between each aligned read and the reference sequence; and (4) genome-wide association study (GWAS), which examines the genomes of different individuals of a species. For each task, we first study the leading computational models based on effectiveness and popularity, and optimize the CPU-based algorithm. Then, we design a new GPU-based parallel algorithm that provides the same interface and functionality as the original CPU-based algorithm. Additionally, we optimize the memory access and disk IO, and schedule the CPU, the GPU, and the IO holistically for efficient co-processing.
Project members

Qiong LUO
Professor
Publications
1. Accelerating Sequence-to-Graph Alignment on Heterogeneous Processors. Zonghao Feng, and Qiong Luo. ICPP 2021: 26:1-26:10.
2. Accelerating Long Read Alignment on Three Processors. Zonghao Feng, Shuang Qiu, Lipeng Wang, and Qiong Luo. ICPP 2019: 71:1-71:10.
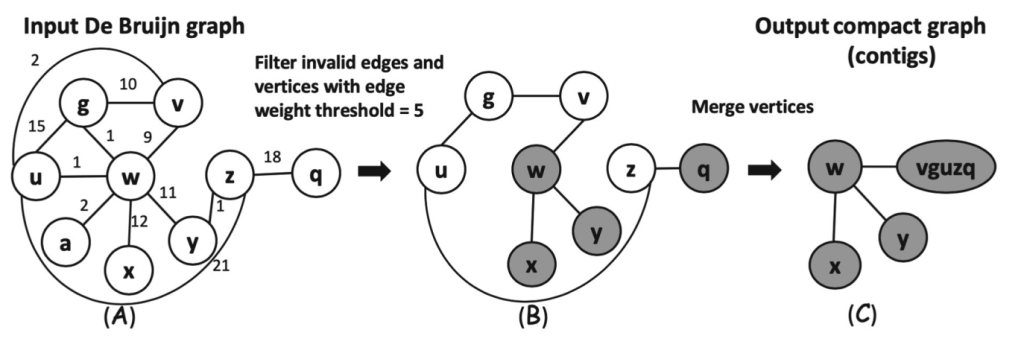
3. Parallelizing Big De Bruijn Graph Traversal for Genome Assembly on GPU Clusters. Shuang Qiu, Zonghao Feng, and Qiong Luo. DASFAA (3) 2019: 466-470.
4. Parallelizing Big De Bruijn Graph Construction on Heterogeneous Processors. Shuang Qiu and Qiong Luo. ICDCS 2017, Atlanta, GA, USA, Jun 2017.
5. Accelerating Large-Scale Genome-Wide Association Studies with Graphics Processors. Mian Lu and Qiong Luo. Wen-Chen Hu and Naima Kaabouch (Editors). IGI Global, 2013. pages 349-380 DOI: 10.4018/978-1-4666-4699-5.ch014.
6. GPU-Accelerated Bidirected De Bruijn Graph Construction for Genome Assembly. Mian Lu, Qiong Luo, Bingqiang Wang, Junkai Wu, and Jiuxin Zhao. APWeb 2013: 51-62.
7. High-performance short sequence alignment with GPU acceleration. Mian Lu, Yuwei Tan, Ge Bai, and Qiong Luo. Distributed and Parallel Databases 30(5-6): 385-399 (2012).
8. Accelerating minor allele frequency computation with graphics processors. Mian Lu, Jiuxin Zhao, Qiong Luo, and Bingqiang Wang. BigMine 2012: 85-92.
9. Integrating GPU-Accelerated Sequence Alignment and SNP Detection for Genome Resequencing Analysis. Mian Lu, Yuwei Tan, Jiuxin Zhao, Ge Bai, and Qiong Luo. The 24th International Conference on Scientific and Statistical Database Management (SSDBM-2012), Chania, Crete, Greece, June 2012.
10. GSNP: A DNA Single-Nucleotide Polymorphism Detection System with GPU Acceleration. Mian Lu, Jiuxin Zhao, Qiong Luo, Bingqiang Wang, Shaohua Fu, and Zhe Lin. The 40th Annual International Conference on Parallel Processing (ICPP-2011), Taiwan, September 2011.
Project Period
2011-Present
Research Area
High-Performance Systems for Data Analytics
